Health
Researchers Uncover How New Genes Emerge and Function in Flies
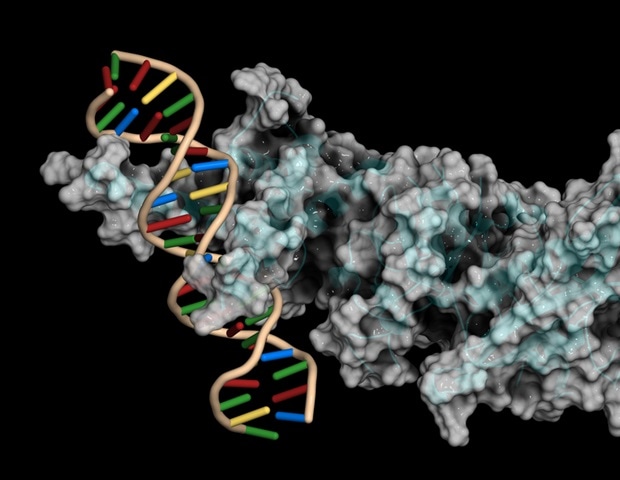
A recent study has revealed significant insights into the regulation of newly emerged genes, known as de novo genes, in fruit flies. After nearly a decade of research, a team from Rockefeller University has documented how these genes are activated and integrated into cellular networks. The findings, published in Nature Ecology & Evolution and PNAS, mark a pivotal step in understanding gene functionality and its implications for evolutionary biology and genetic disorders.
De novo genes are relatively new and originate from previously non-coding regions of DNA. While most genes are ancient and conserved across many species, a small fraction arises spontaneously. According to Li Zhao, Head of the Laboratory of Evolutionary Genetics and Genomics at Rockefeller, understanding how these genes become functional sheds light on broader biological processes, including those involved in diseases such as cancer.
Exploring Gene Regulation
When Zhao established her laboratory eight years ago, the existence of de novo genes was a recent discovery. Over time, she identified hundreds of these genes, prompting Nobel laureate Torsten Weisel to inquire about their regulation during a lunch conversation. This question sparked a long-term investigation into how these genes are expressed.
Recent technological advancements, particularly in computational methods and single-cell sequencing, have enabled Zhao’s team to identify which transcription factors—proteins that help turn genes on or off—regulate specific de novo genes. Their research revealed that only about 10 percent of transcription factors are responsible for the majority of de novo gene expression. By engineering fruit flies with varying levels of these transcription factors and conducting RNA sequencing, the team confirmed their crucial role in gene regulation.
Co-Regulation and Evolutionary Insights
In their study published in PNAS, the researchers examined the genomic neighborhoods surrounding de novo genes to understand their co-regulation with more established genes. By analyzing expression patterns and chromatin accessibility, they found that de novo genes often share regulatory elements with adjacent genes. This suggests a collaborative mechanism in gene regulation, where the cellular environment influences the functionality of these young genes.
Zhao noted the interconnected nature of their findings: one study focuses on how cellular environments regulate new genes, while the other investigates the cooperative operation of genes within the genome. “The papers are closely linked,” she stated, emphasizing the importance of both studies in understanding gene dynamics.
The research also provides insights into the potential origins of de novo genes. Although Zhao cautions that it cannot be definitively stated that transcription factors cause the emergence of these genes, the findings indicate that modifications to these regulatory proteins can lead to significant changes in gene expression.
As Zhao’s lab continues to explore the role of transcription factors in de novo gene regulation, there is hope for deeper understanding of gene network evolution. The implications extend beyond basic biology; they may provide critical insights into diseases characterized by rapid genetic dysregulation, including various forms of cancer.
The ongoing study of de novo genes may also simplify the complex web of gene expression and regulation. Zhao suggests that the relatively straightforward regulation of these young genes could serve as a model for unraveling the intricacies of the entire genome. “Expression and regulation is more complex than we think,” she concluded, highlighting the potential for de novo genes to illuminate fundamental biological questions.
These findings not only advance the understanding of gene regulation but also reinforce the significance of evolutionary biology in addressing human health issues. As research progresses, the connections between de novo genes and broader genetic networks will likely continue to unfold, offering valuable insights into both evolution and disease.
-

 Entertainment3 months ago
Entertainment3 months agoAnn Ming Reflects on ITV’s ‘I Fought the Law’ Drama
-

 Entertainment4 months ago
Entertainment4 months agoKate Garraway Sells £2 Million Home Amid Financial Struggles
-

 Health3 months ago
Health3 months agoKatie Price Faces New Health Concerns After Cancer Symptoms Resurface
-

 Entertainment3 months ago
Entertainment3 months agoCoronation Street’s Carl Webster Faces Trouble with New Affairs
-

 Entertainment3 months ago
Entertainment3 months agoWhere is Tinder Swindler Simon Leviev? Latest Updates Revealed
-

 Entertainment4 months ago
Entertainment4 months agoMarkiplier Addresses AI Controversy During Livestream Response
-

 Science1 month ago
Science1 month agoBrian Cox Addresses Claims of Alien Probe in 3I/ATLAS Discovery
-

 Entertainment4 months ago
Entertainment4 months agoKim Cattrall Posts Cryptic Message After HBO’s Sequel Cancellation
-

 Entertainment3 months ago
Entertainment3 months agoOlivia Attwood Opens Up About Fallout with Former Best Friend
-

 Entertainment6 days ago
Entertainment6 days agoCoronation Street Fans React as Todd Faces Heartbreaking Choice
-

 Entertainment3 months ago
Entertainment3 months agoMasterChef Faces Turmoil as Tom Kerridge Withdraws from Hosting Role
-

 Entertainment4 months ago
Entertainment4 months agoSpeculation Surrounds Home and Away as Cast Departures Mount